- Translation involves “decoding” a messenger RNA (mRNA)
- The translation using its information to build a polypeptide or chain of amino acids.
- A polypeptide is basically a protein with the technical difference being that some large proteins are made up of several polypeptide chains.
The Genetic Code
- Triple : a sequence of three base (a codon ) is needed to specify one amino acid
- No overlapping code : no base are shared between consecutive codon
- Continuous code
- Degenerate : more than one codon can code for the same amino acid
- Universal : same in all organisms
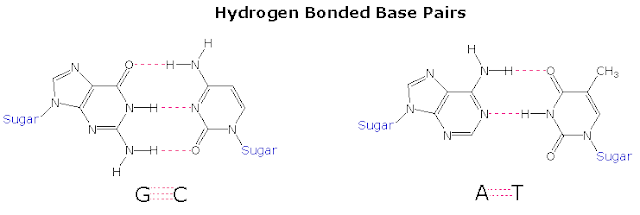
- In an mRNA, the instructions for building a polypeptide come in groups of three nucleotides called codons.
- Have 64 codon
- There are 61 different codons for amino acids
- One codon, AUG, is a “start” signal to kick off translation it also specifies the amino acid methionine. (anticodon for AUG is UAC)
- Three codon ( UAA , UAG and UGA ) “stop” codons mark the polypeptide as finished. (don’t have anticodon).
Translation Requirement
- mRNA (codon) , G C C
- tRNA (anticodon), C G G
- rRNA (ribosome)
- various proteion factors
Step Of Translation The Genetic Message
Step 1 : amino acid activation
- In translation, the codons of an mRNA are read in order from the 5' end to the 3' end by molecules called transfer RNAs (tRNA).
- tRNA has an anticodon, a set of three nucleotides that binds to a matching mRNA codon through base pairing.
- tRNAs bind to mRNAs inside of a protein-and-RNA structure (ribosome).
- As tRNAs enter slots in the ribosome and bind to codons, their amino acids are linked to the growing polypeptide chain (Met-Ile-Ser) in a chemical reaction.
- The end result is a polypeptide whose amino acid sequence mirrors the sequence of codons in the mRNA.
- Free energy of hydrolysis of ATP provides energy for bond formation
- In all organisms , synthesis of polypeptide chain starts at the N- terminal end and grows to C- terminus.
- Initiation required: tRNAfmet ,initiation codon (AUG) of mRNA ,30S ribosomal subunit,50 ribosomal subunit,initiation factors (Ifs) and GTP,Mg2+
- A ribosome (which comes in two pieces, large and small)
- An mRNA with instructions for the protein we'll build
- An "initiator" tRNA carrying the first amino acid in the protein, which is almost always methionine (Met)
- During initiation, these pieces must come together in just the right way .They form the initiation complex.
- Inside our cells and the cells of other eukaryotes, translation initiation , the tRNA carrying methionine attaches to the small ribosomal subunit.
- Together, they bind to the 5' end of the mRNA by recognizing the 5' GTP cap (added during processing in the nucleus).
- Then, they "walk" along the mRNA in the 3' direction, stopping when they reach the start codon (often, but not always, the first AUG).
- Example : tRNAfmet contain the triple 3’-UAG-5’and the triple base pairs with 5’-AUG-3’ in mRNA.
- 3’-UAC-5’ triplet on tRNAfmet recognizes the AUG triplet (the start signal) when it occurs at the beginning of the mRNA sequence that direct polypeptide synthesis (bind at P site).
Eukaryotic translation initiation
Bacterial Translation Initiation
- In bacteria, the situation is a little different. Here, the small ribosomal subunit doesn't start at the 5' end of the mRNA and travel toward the 3' end.
- Instead, it attaches directly to certain sequences in the mRNA.
- These Shine-Dalgarno sequences come just before start codons and "point them out" to the ribosome.
Step 3: Chain Elongation
- Elongation is when the polypeptide chain gets longer.
- Uses 3 biding sites for tRNA present on the 50S subunit of the 70S ribosome: P (peptidyl) site, A(aminoacyl) site ,E (exit) site.
- Requires: 70S ribosome, codons of mRNA , aminoacyl-tRNAs, elongation factors (EF),GTP and Mg2+
- First : a fresh codon is exposed in another slot, called the A site.
: The A site will be the "landing site" for the next tRNA, one whose anticodon is a perfect
(complementary) match for the exposed codon.
:an aminoacyl-tRNA is bound to the A site
: the P site is already occupied by tRNAfmet
: 2nd amino acid bound to 70S initiation complex.Defined by the mRNA.
Second : the peptide bond is formed ,the P site is uncharged.
Third : the uncharged tRNA is released (E site)
: the next peptidyl-tRNA is translocated to the P site
: the next aminoacyl-tRNA occupies the empty A site
- Translation read the mRNA from 5’ to 3’ direction
- Ribosom moves toward 3-end
- Polypeptide sequence grows from N-end to C-end
Step 4 : Chain termination
- Termination happens when a stop codon in the mRNA (UAA, UAG, or UGA) enters the A site.Stop codons have no tRNA,(anticodon).
- Stop codons are recognized by proteins called release factors, which fit neatly into the P site (though they aren't tRNAs).
- Release factor (RFs) which either binds to UAA and UAG or UGA.
- GTP which is bound to RF
- The entire complex dissociates setting free the complete polypeptide ,the release factors,tRNA mRNA ,and the 30S and 50S ribosomal subunits.